Survo is a general environment for statistical computing and related
areas (Mustonen 1992). Its current version SURVO 98 works on standard
PC's and it can be used in Windows, for example. The first versions of
Survo were created already in 1960's. The current version is based on
principles introduced in 1979 (Mustonen 1980, 1982).
According to these principles, all tasks in Survo are handled by a
general text editor. Thus the user types text and numbers in an edit
field which is a rectangular working area always partially visible in a
window on the computer screen. Information from other sources - like
arrays of numbers or textual paragraphs - can also be transported to the
edit field.
Although edit fields can be very large giving space for big matrices
and data sets, it is typical that such items are kept as separate
objects in their own files in compressed form and they are referred to
by their names from the edit field. Data and text are processed by
activating various commands which may be typed on any line in the edit
field. The results of these commands appear in selected places in the
edit field and/or in output files. Each piece of results can be modified
and used as input for new commands and operations.
This interplay between the user and the computer is called
`working in editorial mode'. Since everything is directly based on text
processing, it is easy to create work schemes with plenty of
descriptions and comments within commands, data, and results. When used
properly, Survo automatically makes a readable document about everything
which is done in one particular edit field or in a set of edit fields.
Edit fields can be easily saved and recalled for subsequent processing.
Survo provides also tools for making new application programs.
Tutorial mode permits recording of Survo sessions as sucros (Survo
macros). A special sucro language for writing sucros for teaching and
expert applications is available. For example, it is possible to pack a
lengthy series of matrix and statistical operations with explanatory
texts in one sucro. Thus stories about how to make statistical analysis
consisting of several steps can be easily told by this technique.
Similarly application programs about various topics are created rapidly
as sucros.
Matrix computations are carried out by the matrix interpreter of
Survo (originated in its current form in 1984 and continuously extended
since then). Each matrix is stored in its own matrix file and matrices
are referred to by their file names. A matrix in Survo is an object
having five attributes:
INV(X'*X)*X'*Y,
A matrix has always an internal name, typically a matrix expression
that tells the `history' of the matrix. The internal name is formed
automatically according to matrix operations which have been used for
computing this particular matrix. Also row and column labels - extremely
valuable for identifying the elements - are inherited in a natural way.
Thus when a matrix is transposed or inverted, row and column labels are
interchanged by the Survo matrix interpreter. Such conventions guarantee
that resulting matrices have names and labels reflecting the true role
of each element. Assume, for example, that for linear regression
analysis we have formed a column vector Y of values of regressand and a
matrix X of regressors. Then the command
MAT B=INV(X'*X)*X'*Y
gives the vector of regression coefficients as a column vector (matrix
file) B with internal name INV(X'*X)*X'*Y and with row labels which
identify regressors and a (single) column label which is the name of the
regressand. The following small example illustrates these features.
|
|
Comments:
Above a part of the current edit field is shown as it appears in a
window during a Survo session. The activated Survo commands appear (just
for illustration here) in inverse mode and results given by Survo on a
gray background.
A small table of data values (loaded to the edit field from a larger
data file) is located on edit lines 8-21. It is labelled as a matrix
WORLD12 (line 8). The data set is saved in a matrix file WORLD12 by
activating the MAT SAVE command on line 23. The vector of dependent
variable Life is extracted from WORLD12 by the command
MAT Y!=WORLD12(*,Life). The `!' character indicates that Y will be also
the internal name of matrix file Y. Similarly the matrix of independent
variables is saved as a matrix file X (line 25). The regression
coefficients are computed by the command on line 27 and loaded into the
edit field by the MAT LOAD command on line 28.
Various statistics related to a linear regression model can be
calculated according to standard formulas as follows:
|
|
The computations like those above are suitable in teaching but not
efficient enough for large scale applications. Of course Survo provides
several means for direct calculations of linear regression analysis. For
example, the REGDIAG operation based on the QR factorization of X gives
in its simplest form the following results:
|
|
All statistical operations of Survo related e.g. to linear models and multivariate analysis give their results both in legible form in the edit field and as matrix files with predetermined names. Then it is possible to continue calculations from these matrices by means of the matrix interpreter and form additional results. When teaching statistical methods it is illuminating to use the matrix interpreter for displaying various details step by step.
Typical statistical systems (like SAS, SPSS, S-Plus) as well as
systems for symbolic and/or numerical computing (like Mathematica, Maple
V, Matlab) include matrix operations as an essential part. There is,
however, variation in how well these operations are linked to the other
functions of the system. As far as I know those systems do not provide
natural ways for keeping track on matrix labels and expressions in the
sense described above.
In statistical systems we usually have symbolic labels for variables
and observations and these labels appear properly in tables of results.
However, when data or results from statistical operations are treated by
matrix operations, those labels are discarded. Thus when studying a
resulting matrix it is often impossible to know what is its meaning or
substance without remembering all the preceding operations which have
produced it.
In these circumstances one of the targets of this paper is to show
the importance and benefits of automatic labelling in matrix
computations by examples done in Survo. Another goal is to demonstrate
the interface typical for Survo and co-operation of a statistical system
and its matrix interpreter. Such important aspects as accuracy, speed,
and other self-evident qualities of numerical algorithms have a minor
role in this presentation.
The main example used for describing the capabilities of the matrix
interpreter deals with a certain subset selection problem. It is quite
possible that this problem has been solved in many ways earlier. So far,
however, I have not found any sources of similar attempts. In any case,
in this context it is sufficient that this problem gives several
opportunities to show how the matrix interpreter and Survo in general
can be employed in research work.
Some months before this workshop Simo Puntanen asked me to implement computation of
the basis of the column space of a given mxn matrix A as a new operation
in Survo. When the rank of A is r, this space should be presented as a
full-rank mxr matrix B. I answered by suggesting two alternative
solutions. Both of them are based on the SVD of A since singular values
provide a reliable basis for rank determination which may be a tricky
task in the presence of roundoff (Golub and Van Loan, 1989, p.245-6).
If the SVD of A is A = UDVT, the simplest solution is to select B =
U1 where U1 is formed of the r first columns of U. I felt, however, that
in certain applications it would be nice to stick to a `representative'
subset of columns of A although such a basis is not orthogonal in a
general case. In particular when A is a statistical data matrix, it is
profitable to eliminate superfluous variables which cause
multicollinearity. The problem is now, how to select the `best' subset
of r columns from n possible ones.
There are plenty of books and papers about subset selection, for
example, in regression analysis, but I think that it is another problem
due to presence of a regressand or dependent variable or other external
information.
With my colleagues I had encountered a similar problem in connection
of factor analysis when searching for a good oblique rotation in early
1960's. Our solution was never published as a paper. It has existed only
as an algorithm implemented in Survo.
This approach when applied to the present task is to find a
maximally orthogonal subset. One way to measure the orthogonality of a
set of vectors is to standardize each of them to unit length and compute
the volume of the simplex spanned by these unit vectors. Thus we could
try to maximize the rxr major determinant of the nxn matrix of cosines
of the angles between columns of A. When n and r are large, the total
number of alternatives C(n,r) is huge and it is not possible to scan
through all combinations. Fortunately there is a simple stepwise
procedure which (crudely described) goes as follows:
det = 0
for i = 1:n
k(1) = i
for j = 1:r
det1 = 0
for h = 1:n
det2 = determinant_of_cosines_from(A(k(1)), ..., A(k(j - 1)), A(h))
if |det2| > det1
det1 = |det2|
k(j) = h
end
end
end
if det1 > det
det = det1
for j = 1:r
s(j) = k(j)
end
end
end
In this stepwise procedure each column in turn is taken as the first
vector and partners to this first one are sought by requiring maximal
orthogonality in each step. Thus when j (j = 1, ..., r - 1) columns have
been selected, the (j + 1)'th one will the column which is as orthogonal
as possible to the subspace spanned by the j previous columns. From
these n stepwise selections naturally the one giving the maximum volume
(determinant value) is the final choice.
To gain optimal performance the determinants occuring in this
algorithm are also to be computed stepwise.
It is not guaranteed that A(s(1)), ..., A(s(r)) is the best
selection among all C(n,r), but according to my experience (see notes
below) it is always reasonably close to the `best' one.
The determinant criterion is, of course, only one possible measure
for orthogonality. In many statistical applications it corresponds to
the generalized variance - determinant of the covariance matrix - which
has certain weaknesses as a measure of total variability (see e.g. Kowal
1971, Mustonen 1997) but those are not critical in this context.
Another alternative could be the condition number of the columnwise
normalized matrix, but since it depends on the singular values, it seems
to be too heavy at least computationally.
The QR factorization with column pivoting and a refined form of it
based on the SVD as suggested by Golub and Van Loan (1989, p.572-) for
finding a "sufficiently independent subset" is an interesting
alternative. At least the QR technique might be closely related to the
determinant criterion, but so far I have had not time for comparisons.
The next numerical example illustrates how Survo is used for
calculations of the column space.
|
|
The efficiency of the stepwise procedure will now be compared with
the best possible one - obtained by exhaustive search over all C(n,m)
alternatives - by simulation.
The proportion of cases where the `right' solution is found depends
on parameters m, n, and A itself. Since the value of the determinant
(absolute value of) D = volume/r! is highly sensitive to r, it seems to
be fair to make a monotone transformation and measure orthogonality of a
set of unit-length vectors by the `mean angle' defined as alpha = arccos(x)
where x is the only root in [0,1] of the equation
[1 + (r - 1)x](1 - x)
r - 1
= D, 0 <= D <= 1.
The left hand side of this equation is the determinant of the rxr
matrix
+- -+
| 1 x x ... x |
| x 1 x ... x |
| x x 1 ... x |
| . . . ... . |
| x x x ... 1 |
+- -+
Ratios of alphas of true solution (from exhaustive search) and
stepwise solution (1000 replicates)
m = 5 n = 30 C(n,m) = 142506 m = 10 n = 20 C(n,m) = 184756
S1 S2 S1 S2
ratio N N ratio N N
1 571 818 1 403 752
0.99 - 1 160 92 0.99 - 1 260 167
0.95 - 0.99 260 89 0.95 - 0.99 335 81
0.90 - 0.95 9 1 0.90 - 0.95 2 0
___________________________________________________________
Total 1000 1000 Total 1000 1000
In tables above column S1 tells how close the stepwise solution has been to the best possible one. There are 57.1 % complete hits in m = 5, n = 30 and 40.3 % in m = 10, n = 20. The `mean angle' has never been less than 95% of the best possible.
I tried also improve the percentage of complete hits by extending the stepwise procedure (results given as column S2) as follows. After stepwise selection i (i = 1, ..., n) leading to vectors, say, A(k(1)),A(k(2)), ..., A(k(r)) the improvement is simply:
ready = no
while ready = no
improvement = no
for j = 1:r
for h = 1:n
if A(h) improves the solution when exchanged with A(k(j))
replace current A(k(j)) by A(h)
improvement = yes
break h loop
end
end
end
if improvement = no
ready = yes
end
end
This extension (which still is very much faster than exhaustive
search) seems to improve the number of complete hits and overall
performance. Practically all alphas were at least 99 % of the optimal.
(See S2 columns above).
Since values of alpha are in the interval [0, pi/2], the MAT #MAXDET
command now available in Survo for selecting the most orthogonal basis,
uses 2*alpha/pi as an index of orthogonality. This index has the range [0,1].
The next snapshots from the edit field show how Survo was employed
for making those simulations.
For these experiments a sucro COLCOMP was created first by letting
the system record execution of one typical replicate and then by editing
this sucro program in the edit field. The final form of sucro COLCOMP
reads as follows:
|
|
I do not go into the details of the sucro program code. More
information can be found in Mustonen (1992) and on the website of Survo
(www.helsinki.fi/survo/q/sucros.html).
When this sucro is activated by the /COLCOMP command (line 55) like
any Survo command, it starts a simulation experiment. In this case m =
10, n = 20, and the number of trials is 1000. The seed number for the
pseudo random number generator in the first replicate is 90001 which
will be incremented by 1 in each trial. The results will be collected in
a text file D10_20.TXT.
While running the sucro program shows following information in the
Survo window:
|
|
The display above shows the situation in the third replicate of 1000
experiments. On line 14 a 10x20 random matrix A is generated by using a
seed number 90003. On line 15 the cosine matrix C is computed. On line
16 the MAT #MAXDET(C,m,S,i) command is activated in fact three times
with values i = 0,1,2 as follows (see also lines 33, 38, and 43 in the
COLCOMP program code):
i = 0: Exhaustive search over all C(20,10) combinations,
i = 1: Original stepwise selection (S1),
i = 2: Improved stepwise selection (S2).
The maximal determinant value and orthogonality index are found in
the internal name of the selection vector S displayed by the MAT S
command (line 18). These values are saved by the COLCOMP sucro in an
internal sucro memory (in case i = 2 as Wd2 and Wi2).
After the three alternatives are computed and saved in the internal
memory, the seed number as well as all determinant and index values are
written by COLCOMP on line 21 and saved as a new line in the text file
D10_20.TXT by the COPY command on line 20.
The total execution time of 1000 trials was about 2 hours on my notebook PC (Pentium II, 366MHz). Practically the whole time was spent for the exhaustive search since there were C(20,10)=184756 alternatives to investigate in each trial.
The results are now in a text (ASCII) file, but this file cannot be
used in statistical operations of Survo as such. It must either be
loaded in to the current edit field:
|
|
or copied into a (binary) Survo data file. The latter alternative is
always better for large data sets and done simply by a FILE SAVE
command:
|
|
The text file D10_20.TXT has been copied (line 71) to a Survo data
file D10_20 in which suitable names and types for variables are chosen
automatically. Thereafter the results obtained in simulation can be
studied with various graphical and statistical tools of Survo.
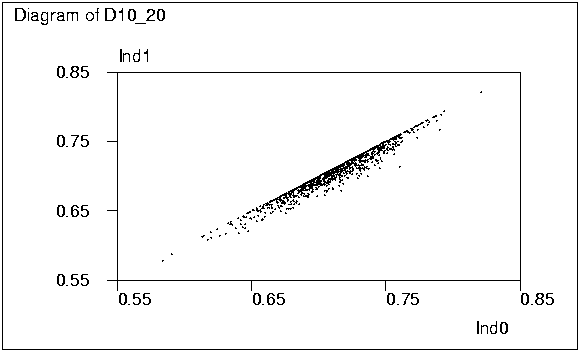
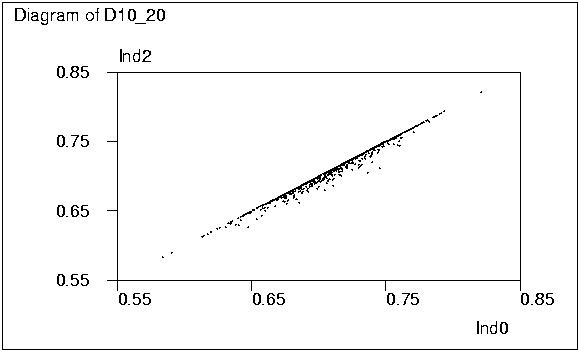
It is interesting to see how closely the two stepwise solutions
Ind1 and Ind2 are related to the `true' one Ind0. The two graphs
obtained by activating the PLOT commands on lines 73 and 74 tell
something about this.
The classified frequencies of ratios Ind1/Ind0 and Ind2/Ind0 given
already as a table were calculated as follows:
|
|
It can also be noted that in this case the distribution of Ind0 is
close to normal while that of Det0 is rather skewed as seen from graphs
plotted by the HISTO commands of Survo.
|
|
The solutions were studied also in higher dimensions. One typical
example is described below.
|
|
This example indicates that only stepwise solutions are possible in
practice. Making of purely random combinations (lines 23-31, option 3 in
#MAXDET) does not produce satisfactory results, but gives a basis for
estimating the time needed for exhaustive search (lines 33-34).
Option 4 (lines 37-43) improves each random combination in the same
way as option 2 improved the original stepwise selection. It seems to
work better than option 3 but is still far away from stepwise
alternatives.
As mentioned earlier the stepwise algorithm has its roots in a
rotation problem of factor analysis. The original idea without explicit
algorithm was presented by Yrjö Ahmavaara in a Finnish text book of
factor analysis already in 1958 with the name Cosine rotation.
When making statistical computer programs I talked about this
problem with Touko Markkanen and Martti Tienari in 1961 and these
discussions led me to the determinant criterion and to the first form of
the stepwise algorithm.
In the standard model of factor analysis (see e.g. Harman, 1967)
with m variables and r common factors the mxm correlation matrix R is
decomposed into the form
(1) R = FphiFT + psi
where F is the mxr matrix of factor loadings, phi is the rxr correlation
matrix of common factors and psi is the mxm diagonal matrix of variances
of unique factors. The decomposition is not unique. If F satisfies (1)
so does also any
(2) A = F(TT)-1
where T is a non-singular rxr `rotation' matrix.
By making suitable assumptions (e.g. multivariate normality of
original variables and by setting extra requirements for F), for
example, a unique ML estimate for F can be found by an efficient
iterative algorithm presented by Jöreskog (1967).
From such a `primary' solution several derived solutions more
pleasant for `interpretation' can be computed by different rotation
methods aiming at `simple structure' so that there are lots of `zeros'
and a suitable number of `high loadings' in the rotated factor matrix A
obtained according to (2).
In the current form of Cosine rotation we try to reach `simple
structure' by selecting the r most orthogonal rows (variables) from F
according to the determinant criterion and letting the r rotated factor
axes coincide with the vectors of these selected variables.
If U is the rxr submatrix of F consisting of those r selected rows
and all columns, the rotation matrix will then be
T = UT[diag(UUT)]-1/2
or in Survo notation simply
MAT T=NRM(U').
Then A will have a structure where for each of the r selected variables
there is one high loading on its own factor and zero loadings in all
other factors.
In order to demonstrate how Cosine rotation works, a simulation
experiment was made as follows. At first a typical F with `simple
structure' and m = 100, r = 7 was selected. Also a suitable factor
correlation matrix phi was given. Then R was computed according to (1),
and a sample of 1000 observations was generated from the multivariate
normal distribution N(0,R) by the Survo command MNSIMUL.
The sample was saved as a Survo data file F100TEST and factor
analysis was performed with 7 factors from this data set. At first
correlations were estimated by the CORR command, then ML solution with 7
factors was found by the FACTA command, and Cosine rotation was
performed by the ROTATE command. Finally the rotated factor matrix A was
compared with the original F.
The following snapshots from the edit field tell the details:
|
|
These MAT commands have produced the original factor matrix F. Most
of the rows are purely random. To introduce some kind of simple
structure, 7 rows are replaced systematically by vectors Y1, ..., Y7
each of them having one high loading while other loadings are pure
zeros. My aim is to show that Cosine rotation is able to detect this
structure from simulated data when the sample size is large enough.
Thus the F matrix has the following contents:
|
|
Let all the correlations between factors be rho = 0.2. Then the factor
correlation matrix phi and the correlation matrix R having the structure
(1) are computed as follows:
|
|
In the next display line 143 tells how a sample of 1000 observations
from the multivariate distribution N(0,R) is generated by an MNSIMUL
command. The second parameter (*) indicates that means and standard
deviations of all variables are 0 and 1, respectively. The sample (100
variables and 1000 observations) is saved in a Survo data file F100TEST.
The sample correlations are computed from this data file and saved
as a 100x100 matrix file CORR.M by the CORR command on line 145.
The maximum likelihood factorization with 7 factors is performed by
the FACTA command (line 146) from the correlation matrix CORR.M. The
100x7 factor matrix is saved as a matrix file FACT.M.
Finally, Cosine rotation is made by the ROTATE command (line 147)
and the rotated matrix is saved as AFACT.M and loaded into the edit
field (lines 148-178; certain lines are deleted afterwards).
Line 149 (giving the labels of the columns) is very important. It
tells that variables Y1, ..., Y7 - which were supposed to be factor
variables - really form the selected, maximally orthogonal basis for the
solution. Already this proves that we have found the original structure
from a noisy data. Here we have again a situation where alphanumeric
labels turn out to be very useful.
|
|
The ROTATE command has saved the rotation matrix T as TFACT.M and
the estimate of factor correlations as RFACT.M. We see that all
correlations (lines 190-198) are reasonably close to the theoretical
value rho = 0.2.
It remains to be checked how good is the match between the original
factor matrix F (saved as matrix file F) and the rotated factor matrix
A = AFACT.M obtained from the sample.
Due to possible ambiguity in rotation it is reasonable to seek an
rxr transformation matrix L which minimizes
f(L) = ||AL - F||2.
The solution of this least squares problem is
L = (ATA)-1ATF.
Besides L - especially in connection with factor analysis - it is
worthwhile to compute the residual matrix
E = AL - F
or compare AL and F by other means as done in general situations by
Ahmavaara already in 1954. Here we only compute the sum of squares of
the elements of E which is the same as the minimum of f(L) squared.
|
|
Matrix L is rather accurately a permuted identity matrix as seen on lines 204-213. Thus there is almost a perfect match between the original and estimated factors. The residual sum of squares is fairly small (line 217) and confirms that this form of factor analysis (in this case) has managed to recover the original factor structure.
It should be noted that if the data set F100TEST is centered and
normalized by columns (variables) and the most orthogonal submatrix is
searched for by the MAT #MAXDET operation (thus applied to CORR.M), none
of the Y variables appear among the 7 selected ones:
|
|
This shows how important it is to purify the data from extra noise caused by unique factors before trying to find a simple structure and this happens by making first the decomposition (1) by ML principle, for example.
It is also interesting to see how many observations are actually
needed in this example for detecting the correct structure.
By a simple sucro similar factor analyses were made for all
subsamples of observations 1, 2,..., N for N = 101, 102,..., 1000 and in
each case the residual sum of squares, maximum absolute residual, and a
dummy variable telling whether the `correct' Y basis was found, were
calculated.
This information was collected in a Survo data file and the
following graphs could then be plotted.
The largest N for which the original basis was not found was 749.
Even in this case six of the seven Y variables was selected but Y5 was
replaced by X55 which was the variable with highest correlation with Y5.
Similar digressions took place also in many other cases, but anyhow when
N was above 300 one can say that the original factor structure was
decently recovered. At least below 250 both the residual sums of squares
and the maximum residuals are so high that finding the original
structure often fails.
To be on the safe side, in this kind of situation at least 600
observations should be available. I think that in too many real
applications which I have seen during the past 40 years, the number of
observations has been too small or the sample has been too
heterogeneous.
For certain, partly justified reasons factor analysis has bad reputation among statisticians. I feel, however, that sometimes the criticism has gone too far or rests on false premises as done by Francis (1974) and quoted by Seber (1985). Some of the erratic conclusions of Francis have been rectified by Mustonen and Vehkalahti (1997).
SURVO 98 is a collection of hundreds of program modules and other system files. All the modules are working under the control of the Survo Editor which is the visible part of the system (as seen in all previous displays) and this editor calls program modules into use as child processes according to the activations of the user. The user sees Survo as one large program or environment without a need to know about the internal structure of the system.
When a command in the edit field is activated, the Editor recognizes and executes it. Simple commands (like those related to text processing or small-scale calculations) are performed by the editor itself. Other commands (like statistical and graphics commands) are only interpreted to certain extent and then the editor calls a suitable module (i.e. spawns it as a child process) for actual execution. The module obtains all pertinent information from the editor (through a single pointer that points at a table of pointers of Survo's internal workspace) and calculates and produces results to be shown in the edit field and/or in matrix files and other files for the results. After termination of the child process the control returns to the editor with eventual results properly displayed and the user can continue the work.
This architecture enables continuous extension of the system itself even while using it. In fact, all programming and system development related to Survo is done while using Survo. Program modules are written in C according to certain simple rules described in Mustonen (1989) and in www.helsinki.fi/survo/c/index.html. For example, a new module is created by typing the program code in the edit field, then saving it, and compiling it by the Watcom C Compiler with the DOS /32A Extender. As a result we have a new Survo module which immediately can be activated by its name like any other module. The modules are genuine 32-bit programs in protected mode and they have access to all memory up to 2 gigabytes in the CPU.
All commands of the matrix interpreter have MAT as their first `word'. When such a command is activated, the editor calls the matrix interpreter (module _MAT.EXE) which executes most MAT commands directly. Certain additional commands (like MAT #MAXDET) are external to the matrix interpreter. They appear in their own program module(s) and are called in turn by the matrix interpreter (as `grandchildren' of the editor).
The matrix interpreter (as other Survo modules) uses dynamic space
allocation for all workspace they need during their use. This space is
freed automatically after execution of the command. For example, in
matrix multiplication (MAT C=A*B) space is reserved for the operands A
and B as well as for the result C just according to their true
dimensions. The operands must be located as matrix files (as A.MAT and
B.MAT in the example). After they are loaded into the central memory the
results are computed and saved as matrix files (C.MAT in the example).
Typically only three spaces for matrices are used simultaneously. Thus
more complicated commands like computing of a Mahalanobis' distance by
MAT D2=(X-MY)'*INV(S)*(X-MY)
are automatically preprocessed and converted to a series of basic matrix
commands. The above MAT command is translated within the matrix
interpreter into 4 MAT commands
MAT %%1=X-MY
MAT %%2=INV(S)
MAT %%2=%%2*%%1
MAT D2=%%1'*%%2
where %%1, %%2,... are temporary matrices (matrix files).
Although a lot of loading and resaving of matrices takes place
during matrix computations, no significant loss is encountered in
execution times due to proper cache memory settings. On the contrary, it
is worthwhile to have all matrices automatically saved in permanent
files even if the work is suddenly interrupted for some reason...
I am grateful to Simo Puntanen and Kimmo Vehkalahti, who have been helpful in many respects during preparation of this manuscript.
The website of Survo (www.helsinki.fi/survo/eindex.html) contains more information about the topic of this talk.
This paper has been prepared entirely by using SURVO 98 and it is
printed by the PRINT operation of Survo on a PostScript printer.
The paper has also been converted into HTML format by Kimmo Vehkalahti
by his own HTML driver for the PRINT operation.